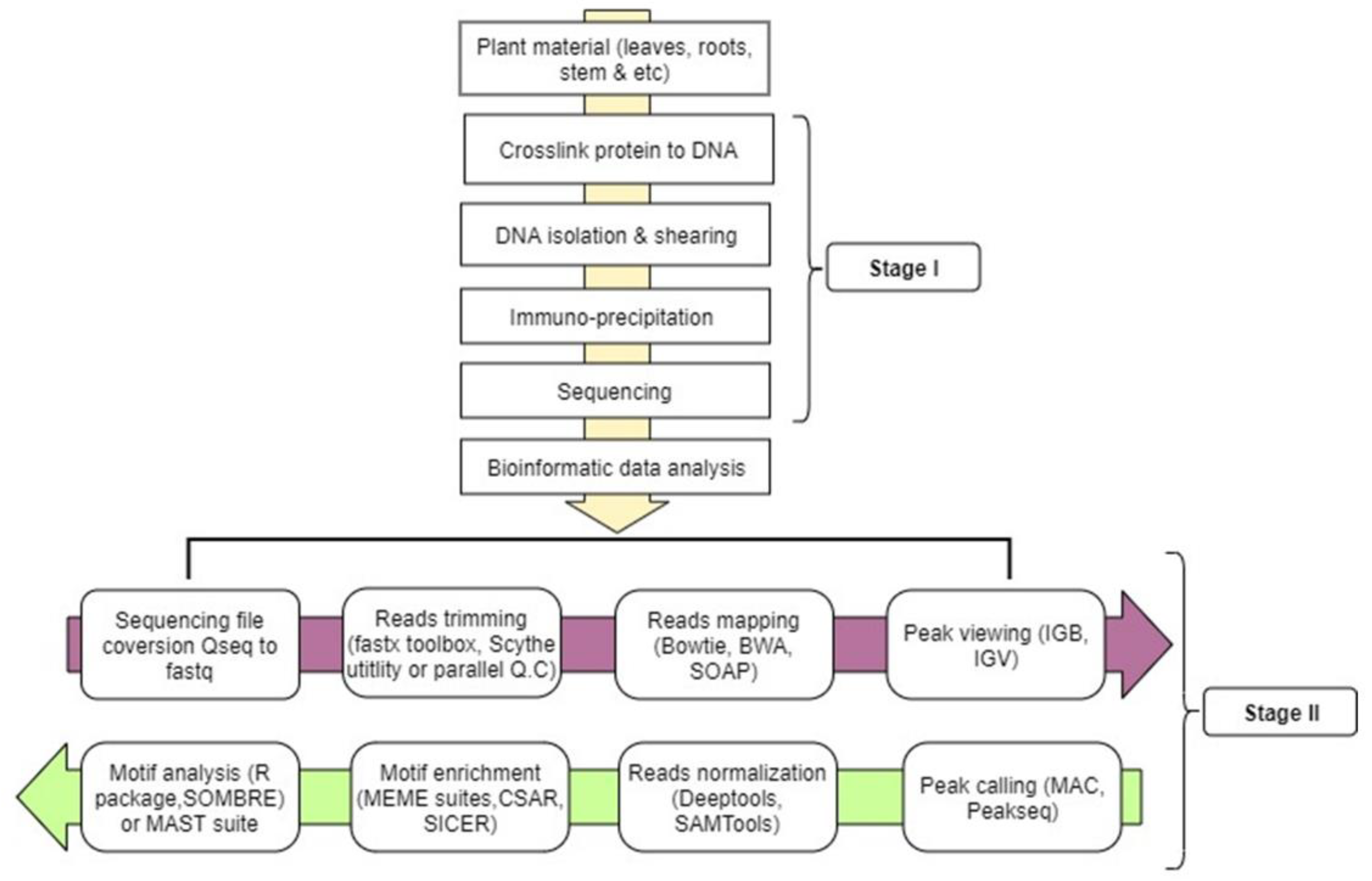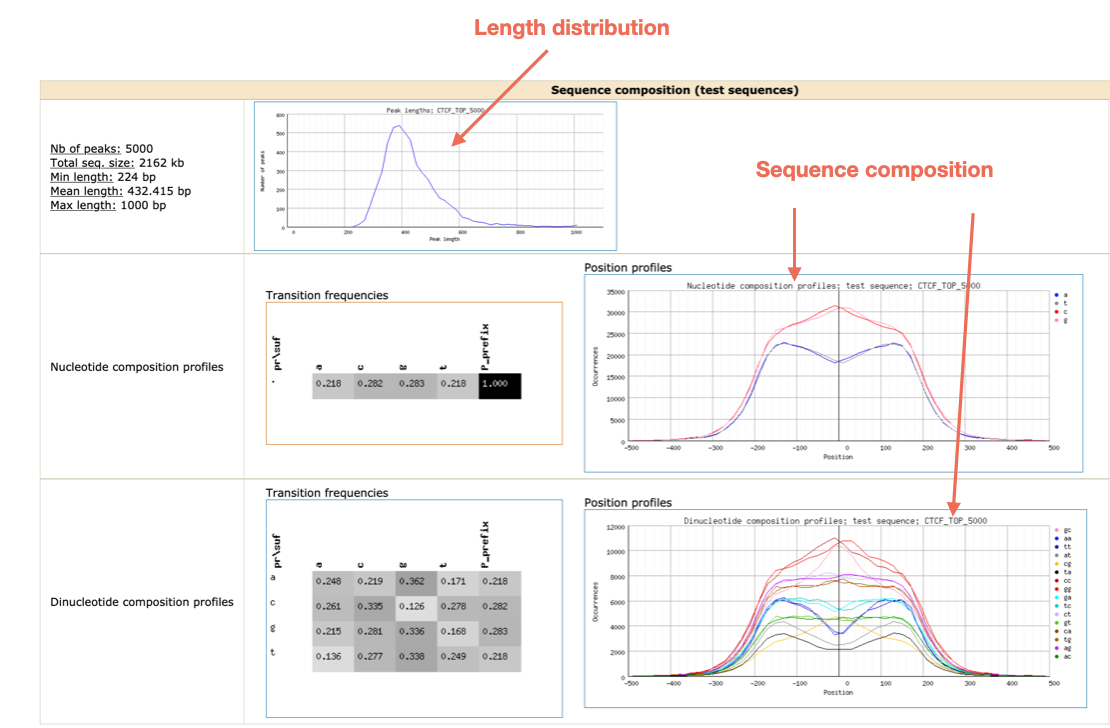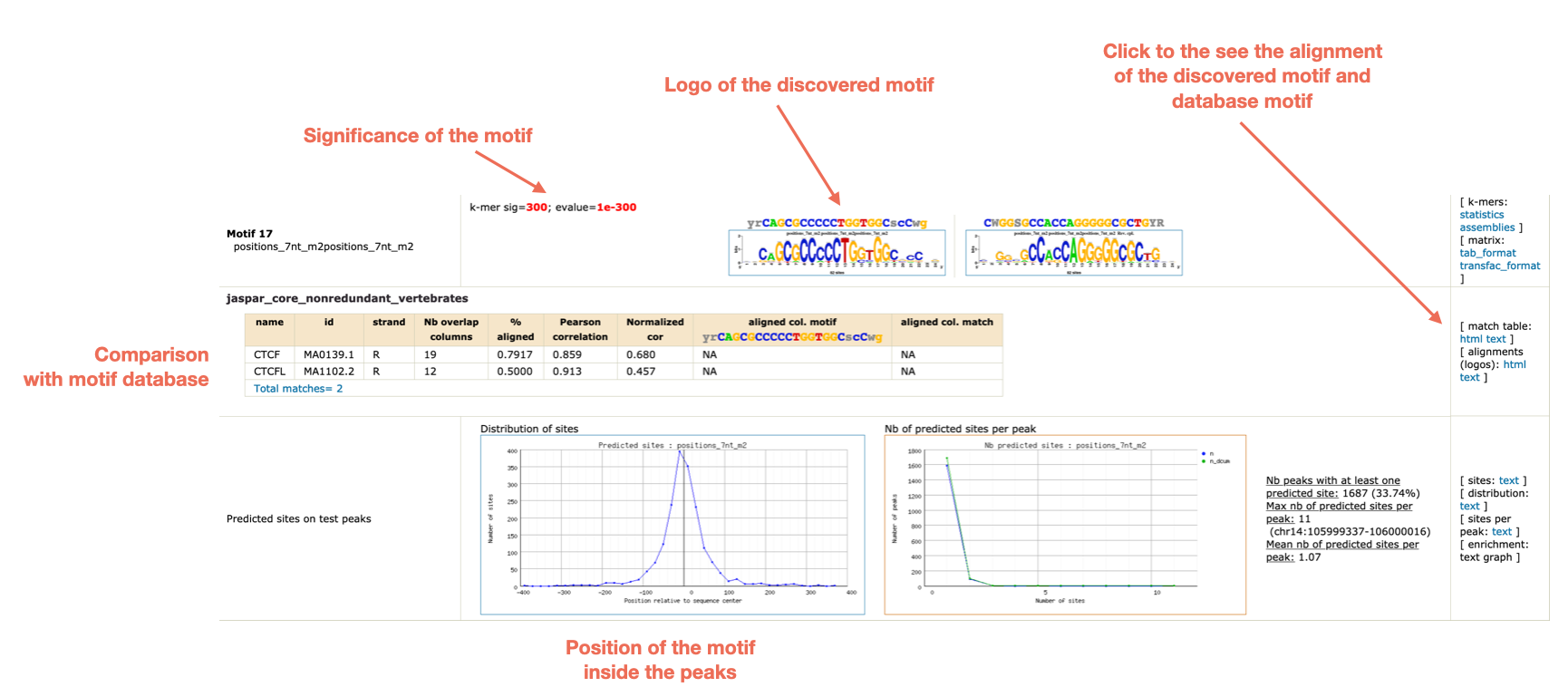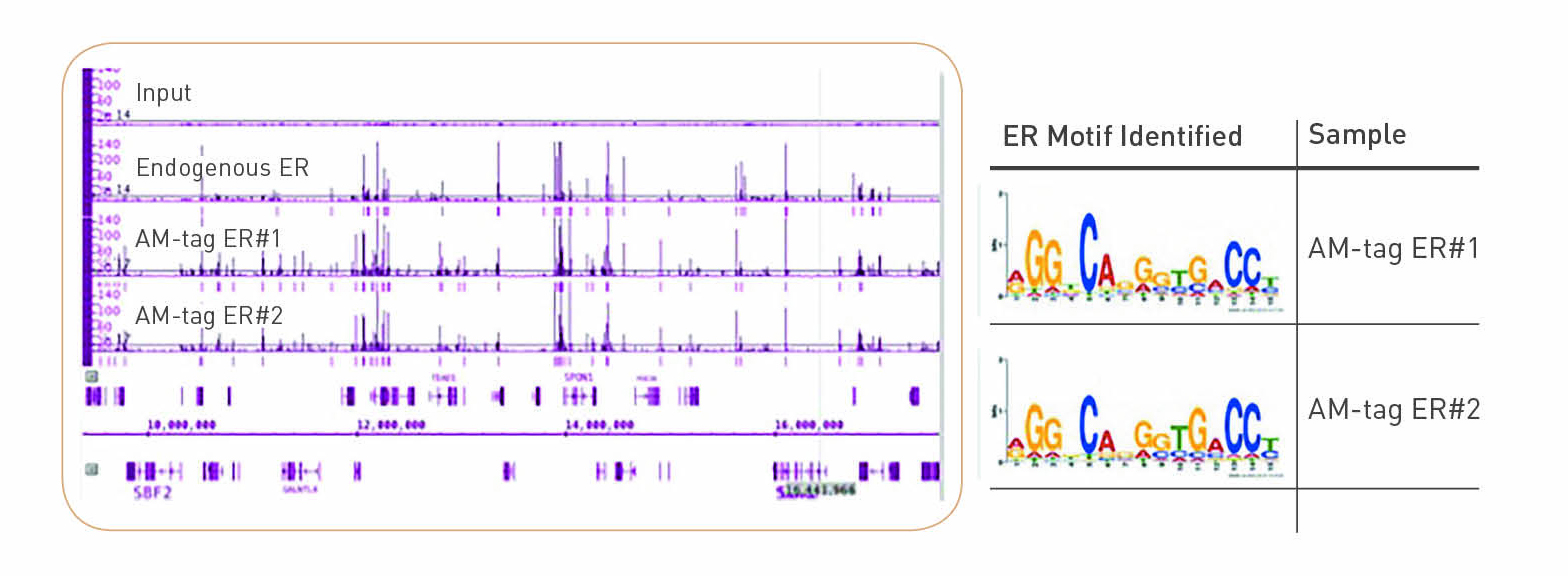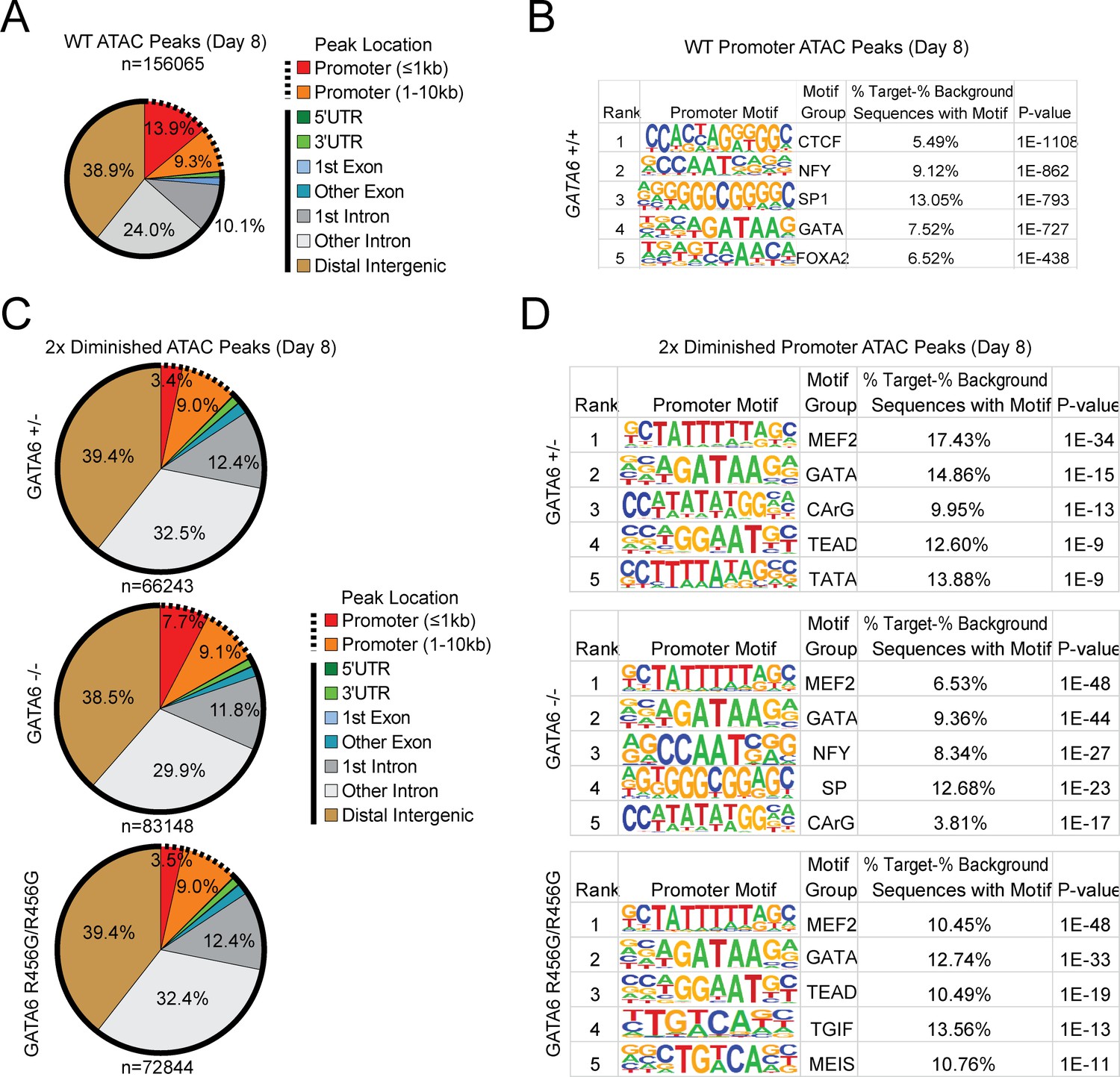
Figures and data in GATA6 mutations in hiPSCs inform mechanisms for maldevelopment of the heart, pancreas, and diaphragm | eLife
NF90/ILF3 is a transcription factor that promotes proliferation over differentiation by hierarchical regulation in K562 erythroleukemia cells | PLOS ONE
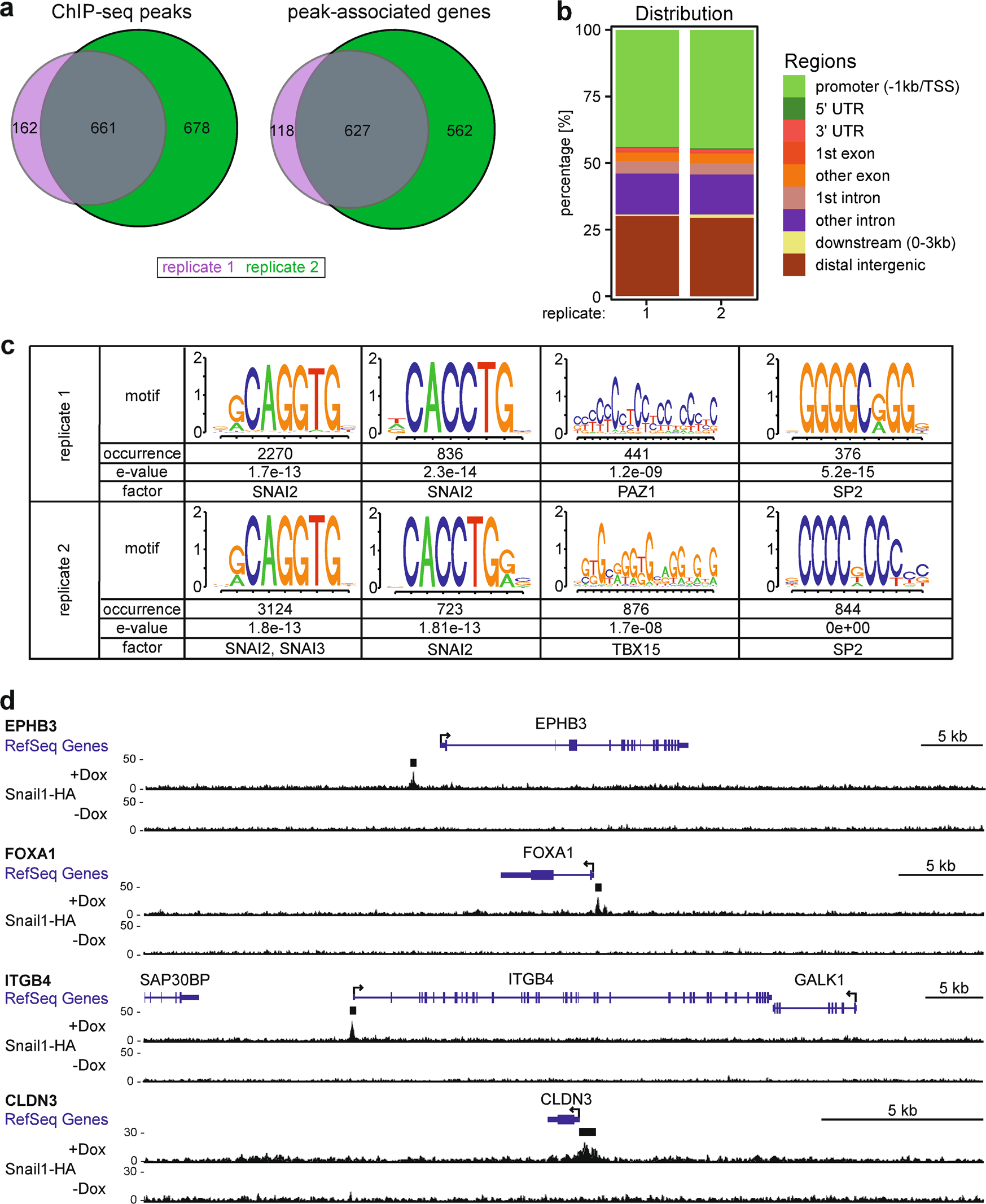
Genome-wide mapping of DNA-binding sites identifies stemness-related genes as directly repressed targets of SNAIL1 in colorectal cancer cells | Oncogene

Motif analysis of TCF4 ChIP-seq data. The signifpeaks.gff file for the... | Download Scientific Diagram

The known motif for the ChIP-ed motif is more enriched in ChIP-seq peak... | Download Scientific Diagram
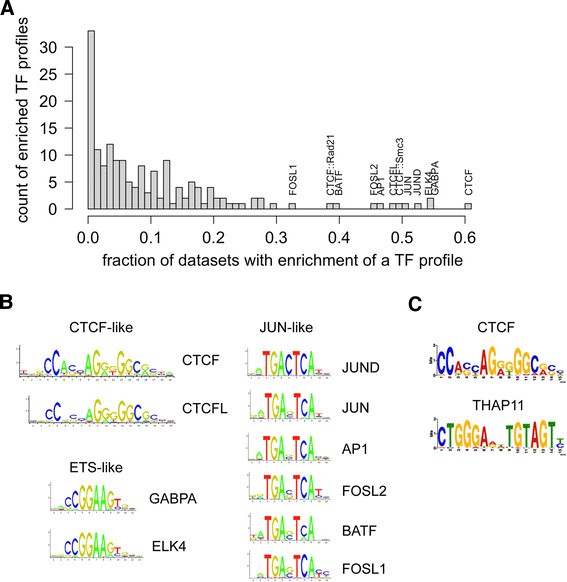
Non-targeted transcription factors motifs are a systemic component of ChIP- seq datasets | Genome Biology | Full Text

JCI - Histone methyltransferase MLL4 controls myofiber identity and muscle performance through MEF2 interaction
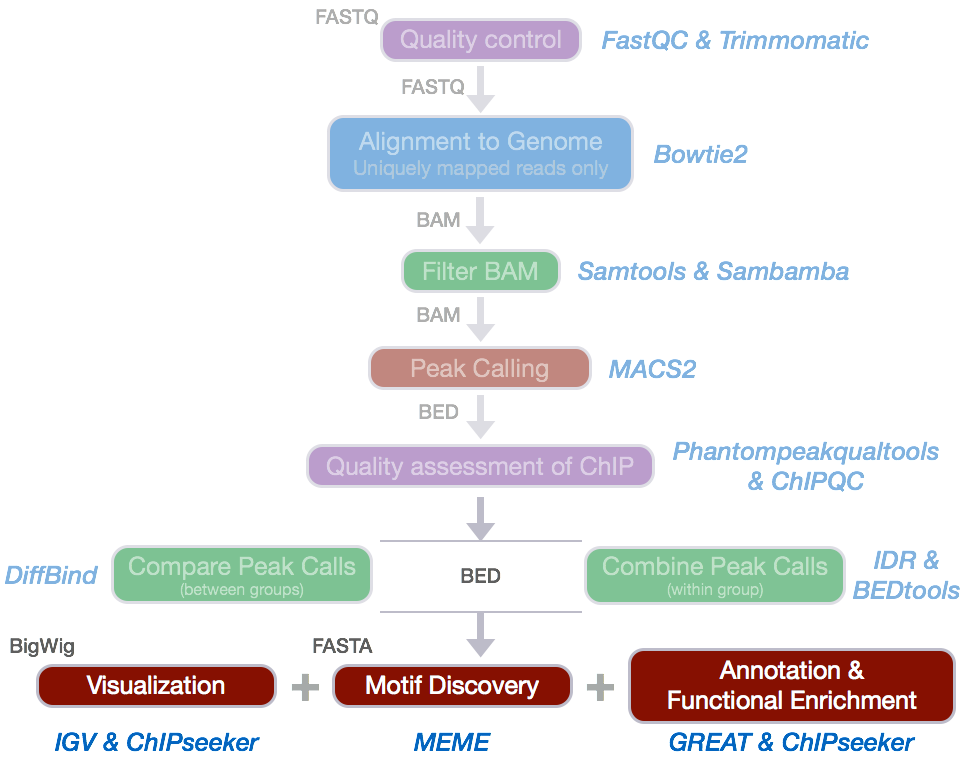
ChIP-seq Peak Annotation and Functional Analysis | Introduction to ChIP-Seq using high-performance computing

Figure 4 from Microfluidic affinity and ChIP-seq analyses converge on a conserved FOXP2-binding motif in chimp and human, which enables the detection of evolutionarily novel targets | Semantic Scholar

Genomic location annotation and motif analysis of SRF ChIP-seq peaks in... | Download Scientific Diagram

Homer motif analysis: "Very close % of target vs % of Background sequences with Motif" & "SeqBias at 1st rank"

Motif identification and analysis a, The 293 high-confidence motifs... | Download Scientific Diagram