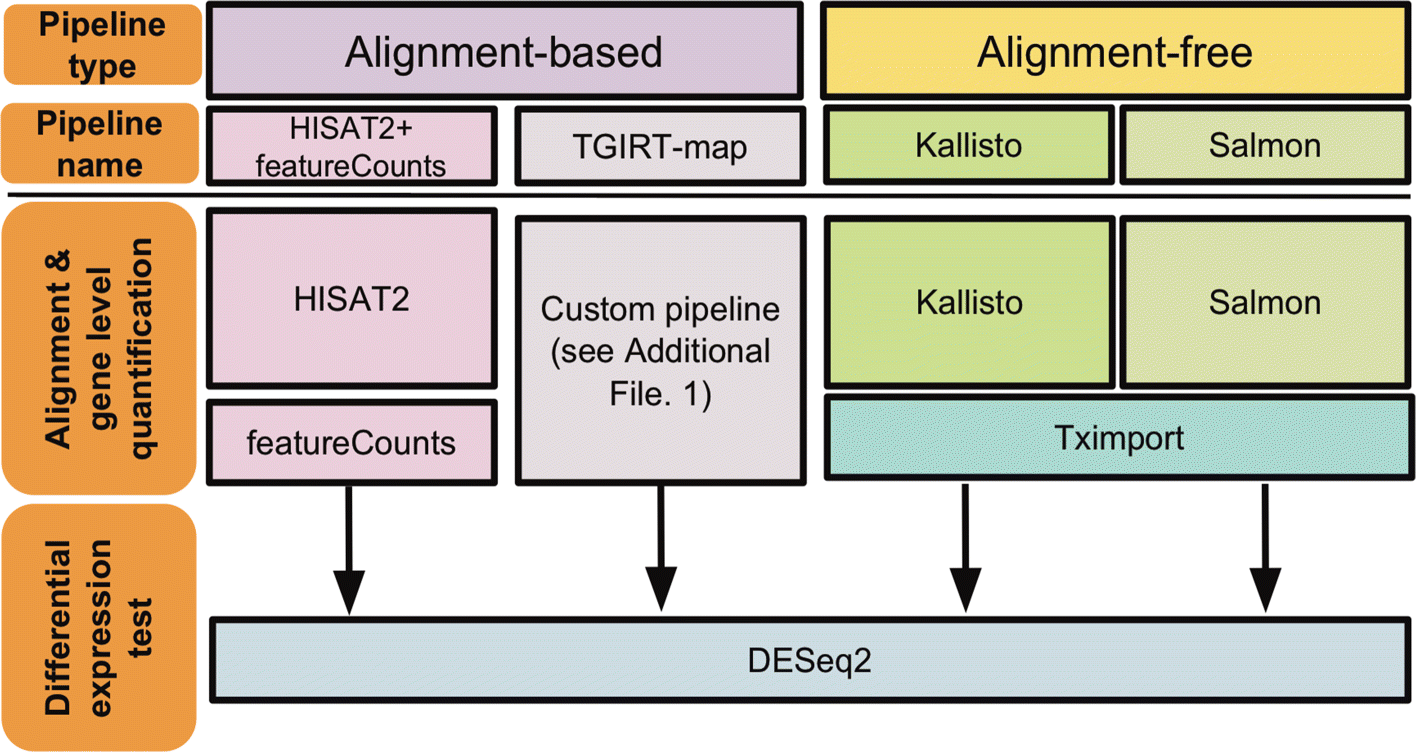
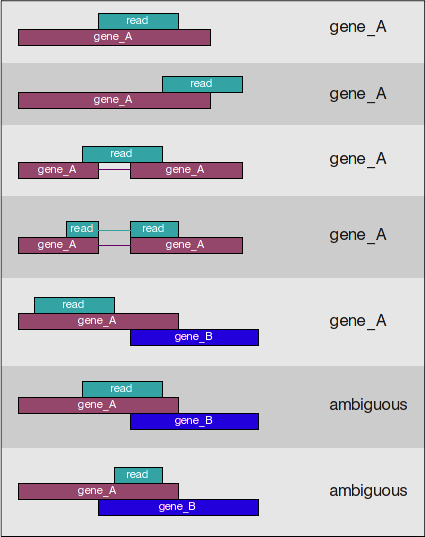
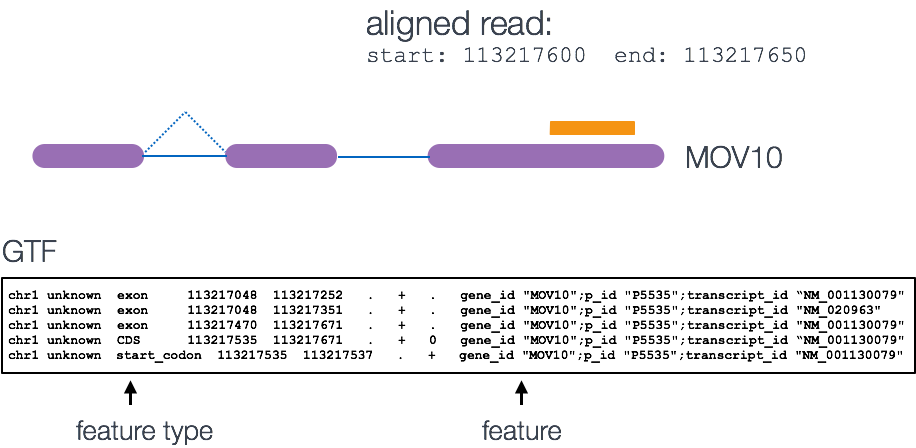
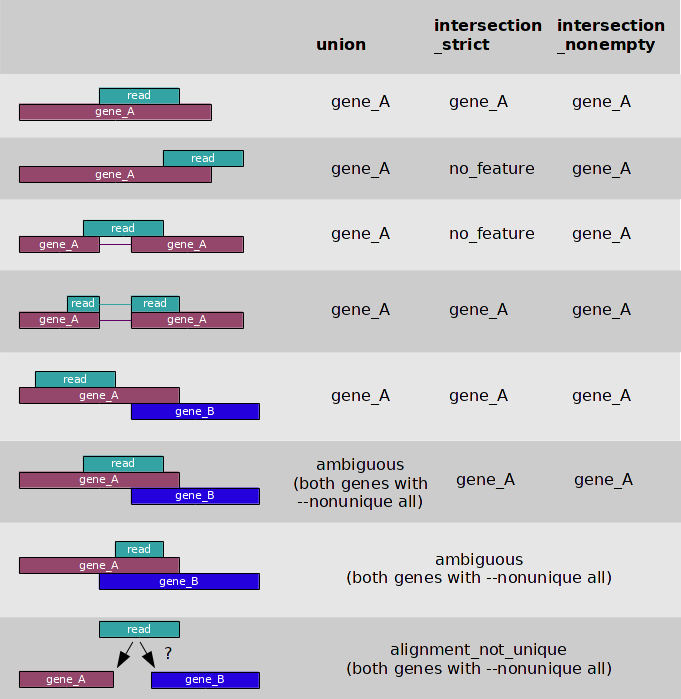
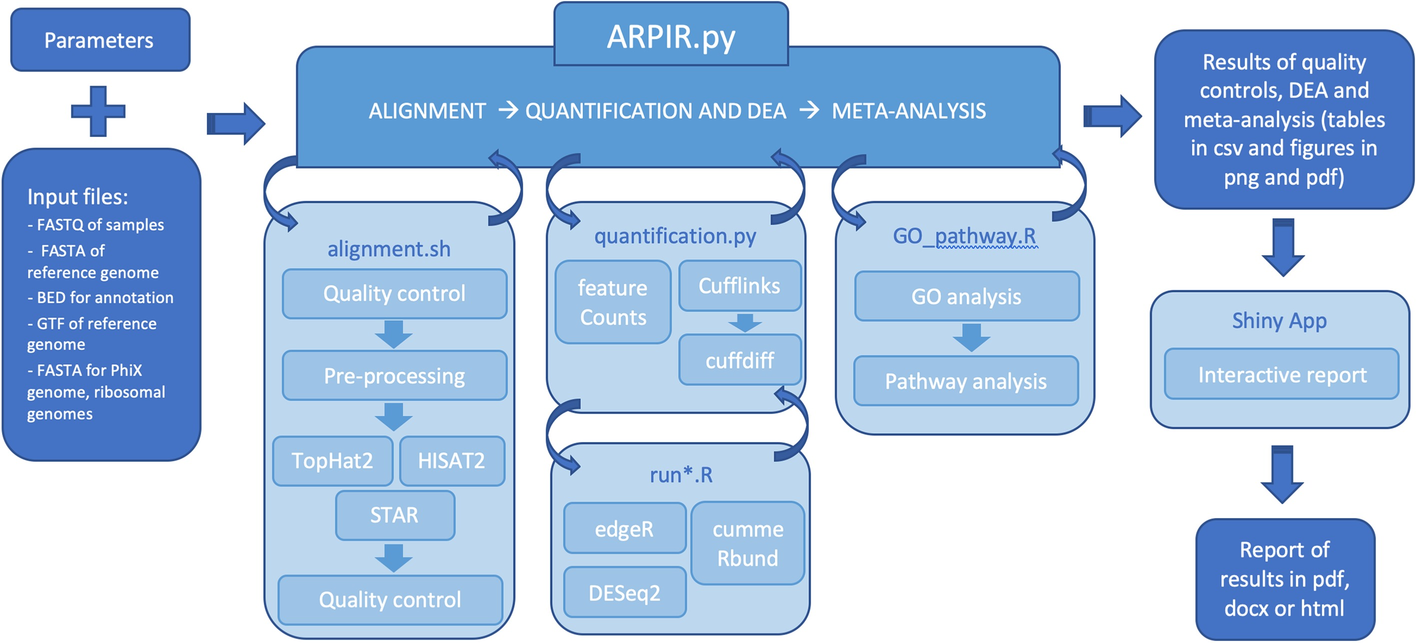
Implementation of FADU. (A) The workflow of FADU uses a BAM file and a... | Download Scientific Diagram
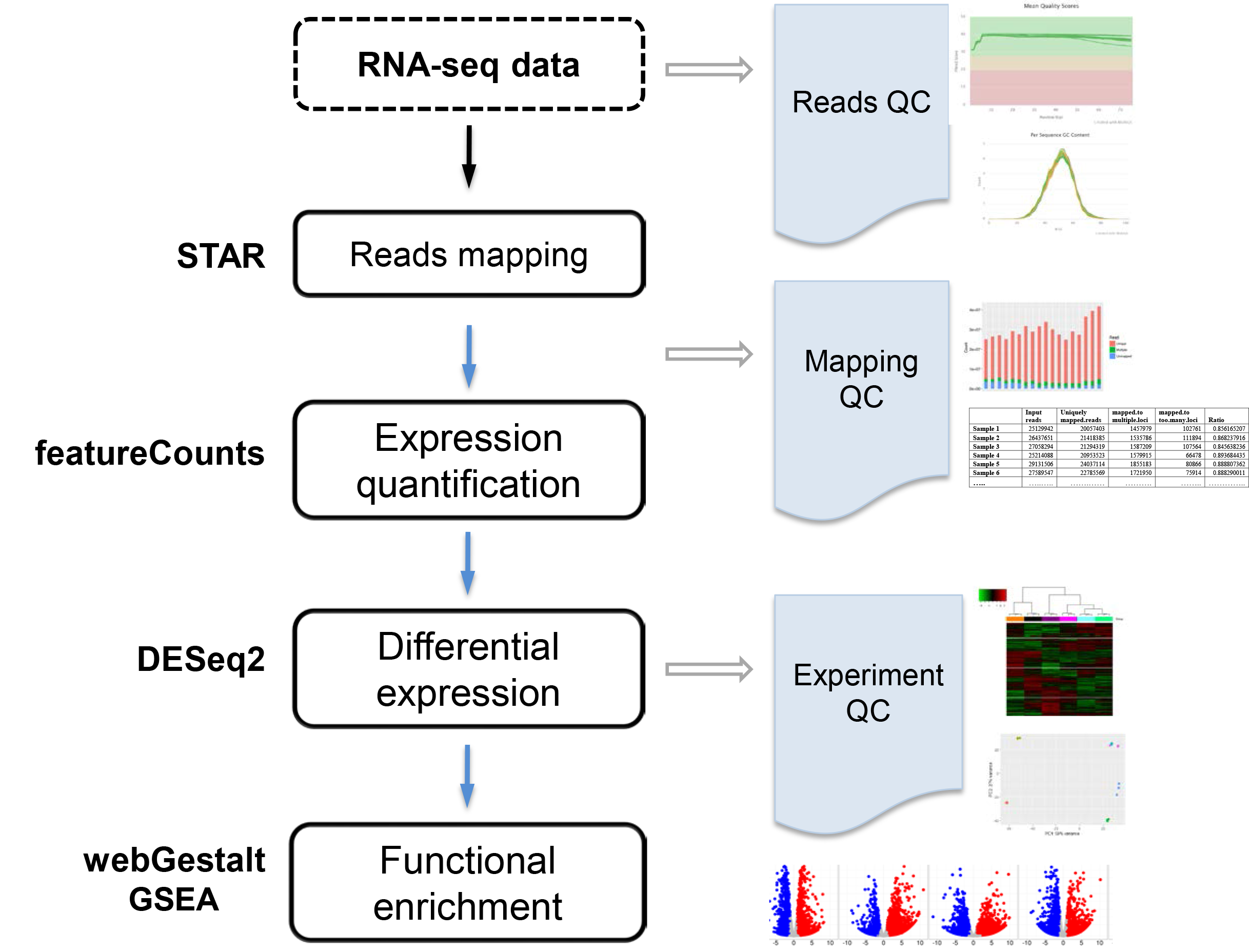
Genes | Free Full-Text | RNAflow: An Effective and Simple RNA-Seq Differential Gene Expression Pipeline Using Nextflow | HTML
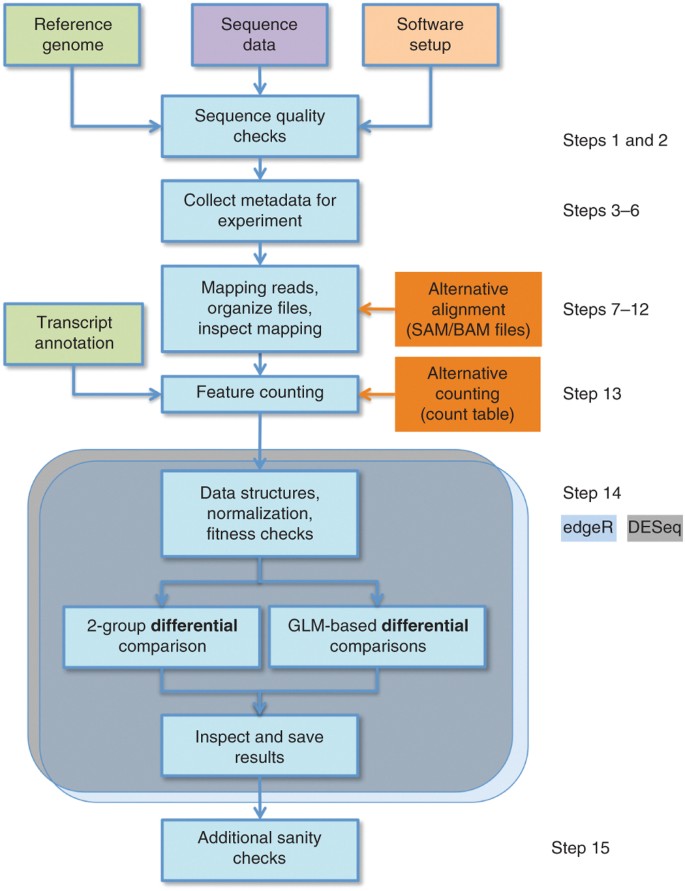
Count-based differential expression analysis of RNA sequencing data using R and Bioconductor | Nature Protocols
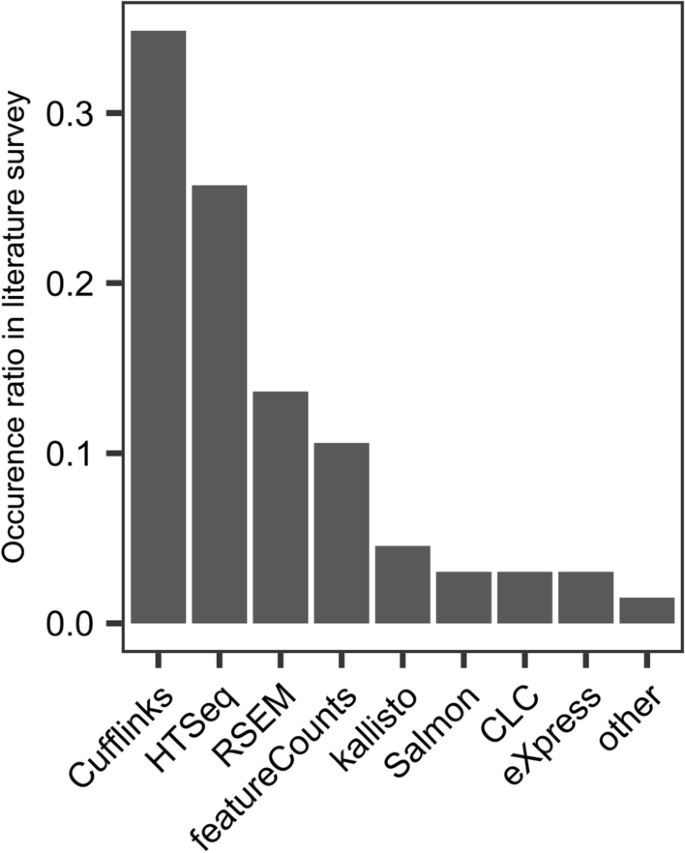
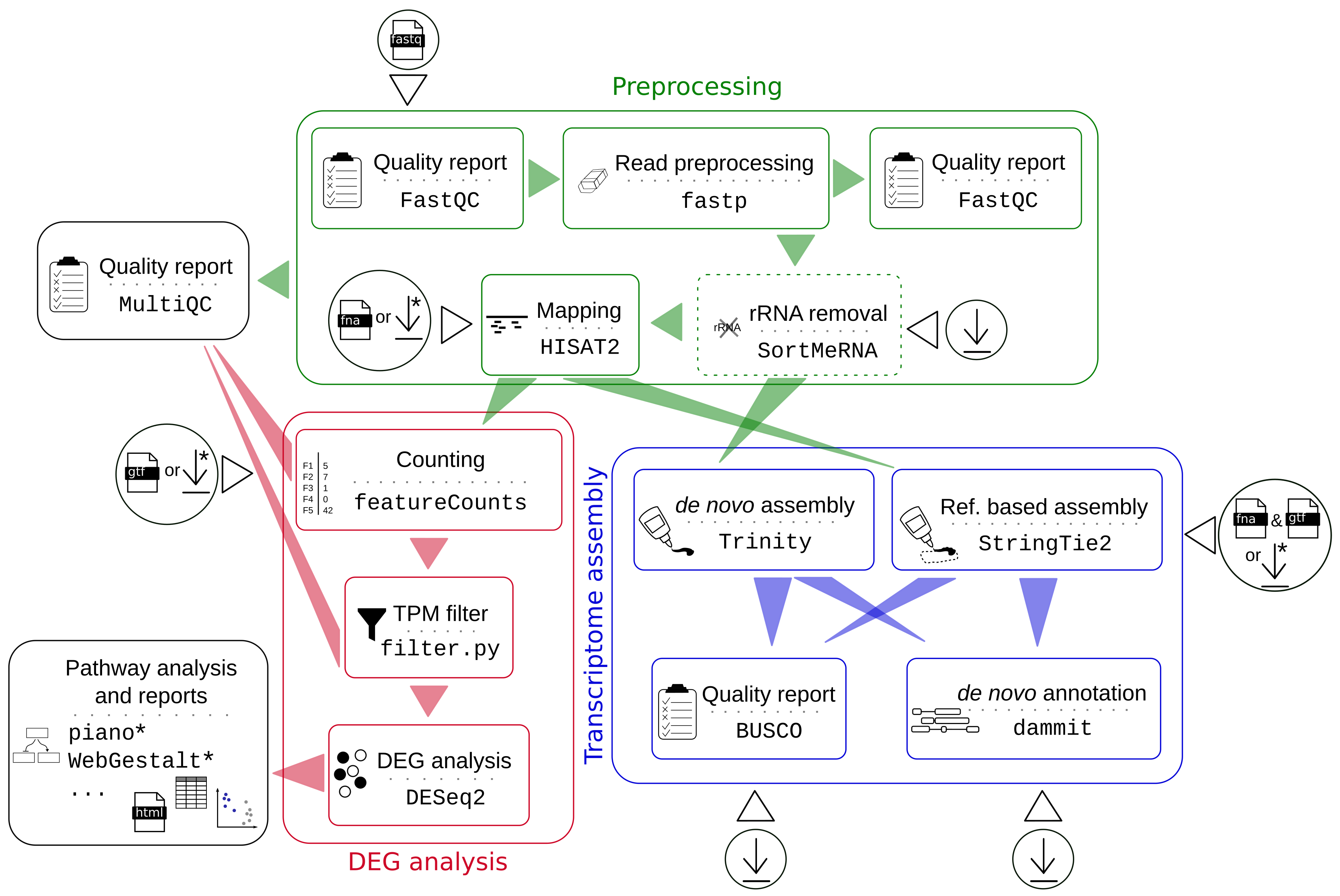
Comparative evaluation of full-length isoform quantification from RNA-Seq | BMC Bioinformatics | Full Text


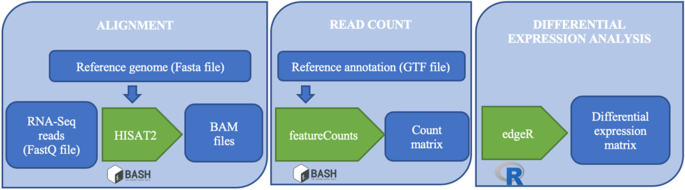
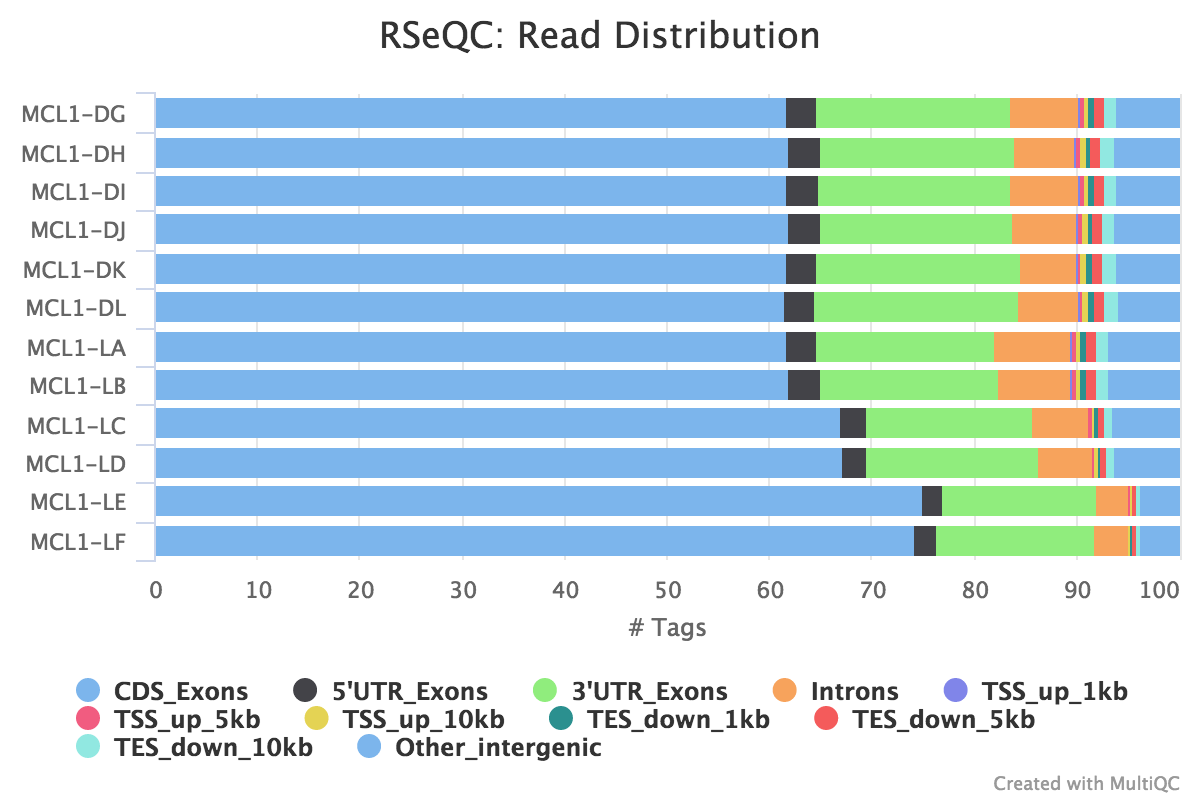
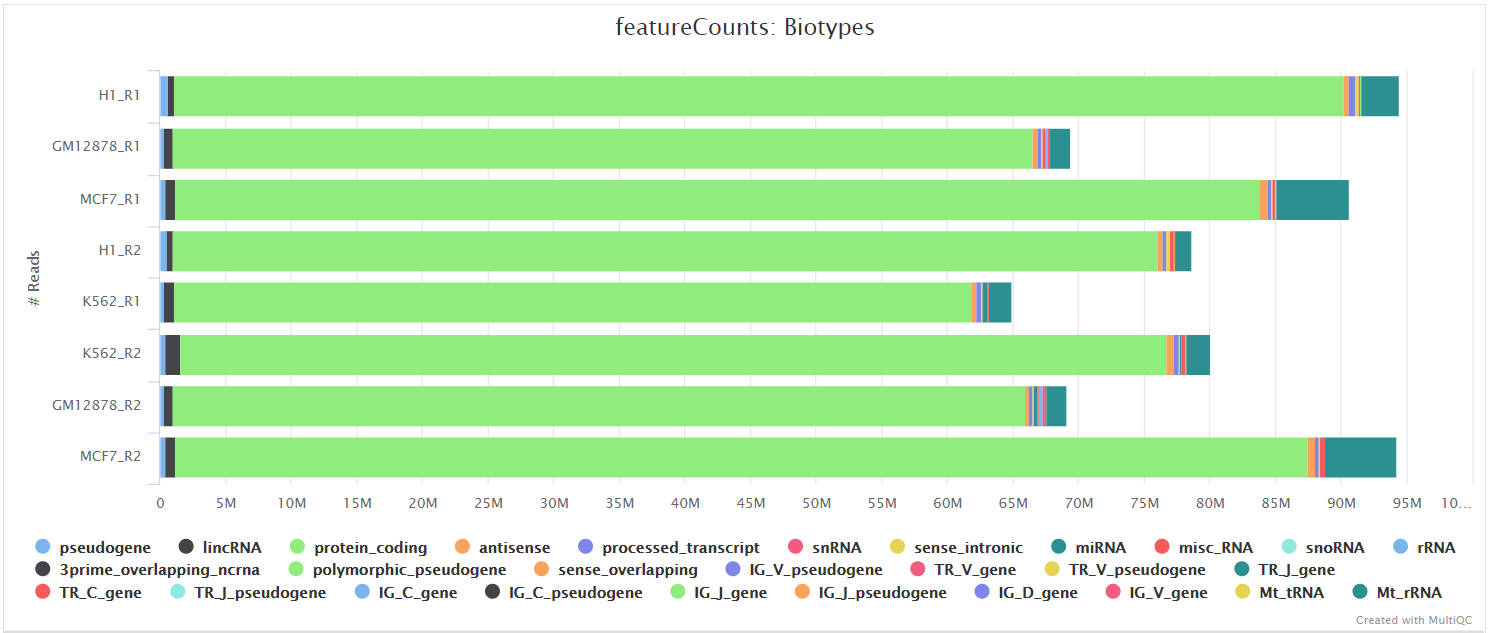

![PDF] Rcount: simple and flexible RNA-Seq read counting | Semantic Scholar PDF] Rcount: simple and flexible RNA-Seq read counting | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/6dc7d659b828d42c1a3d9d968824c43123b19ac2/2-Figure1-1.png)