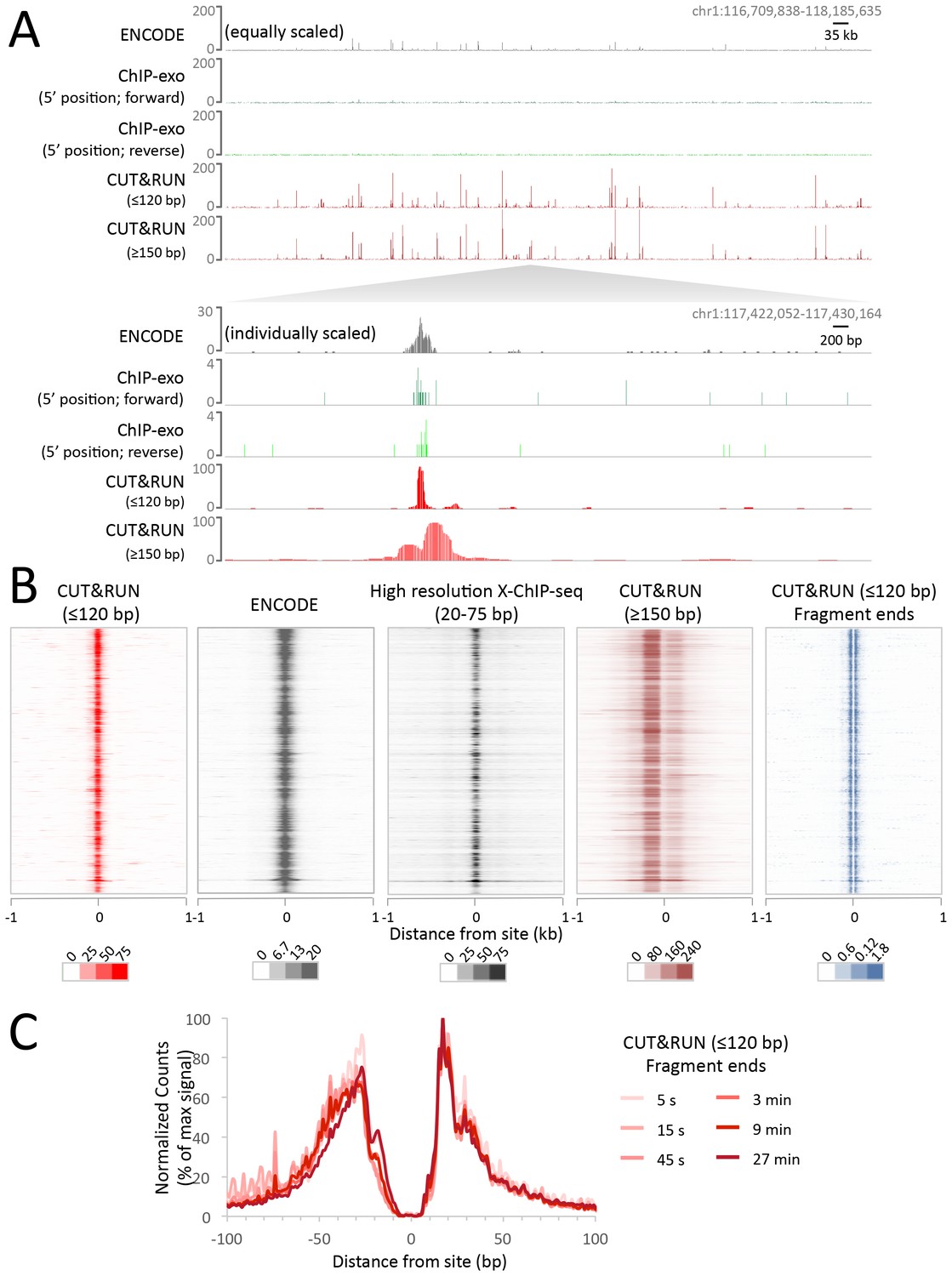
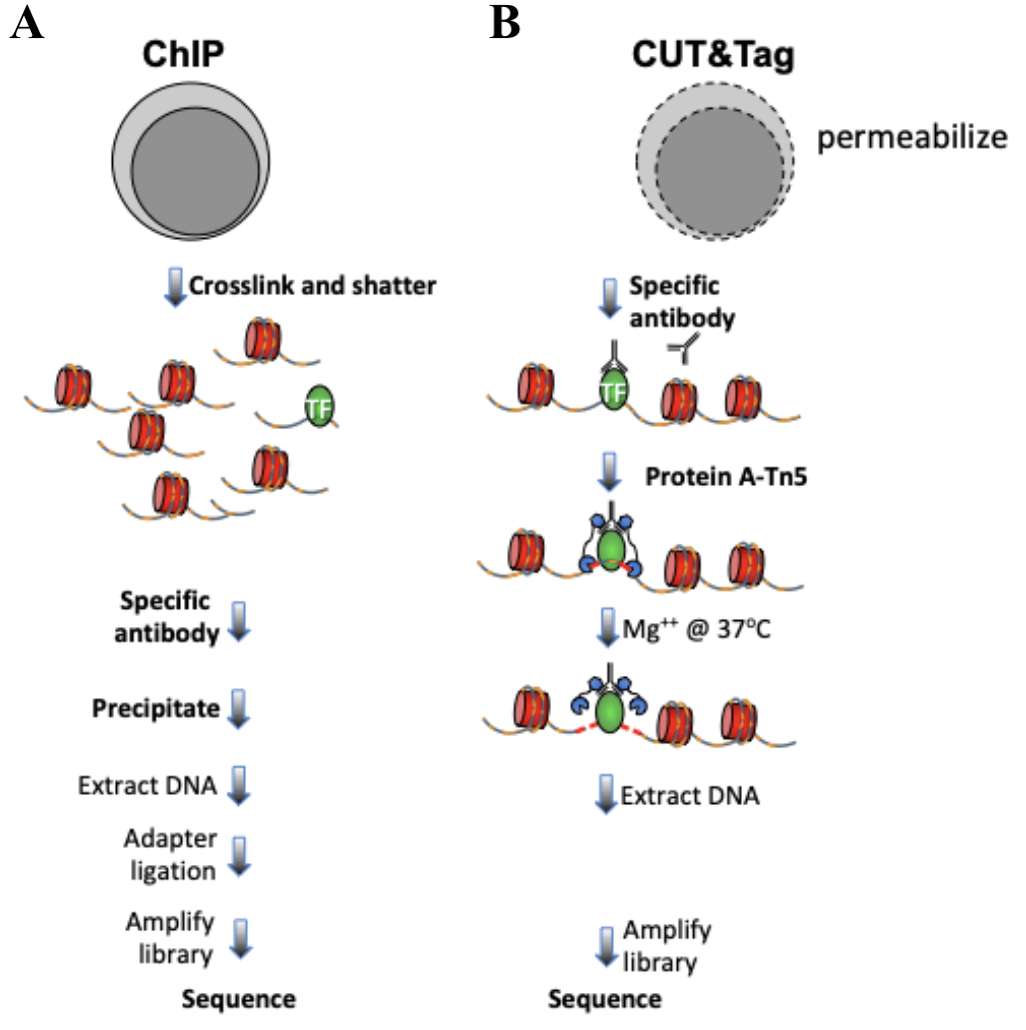
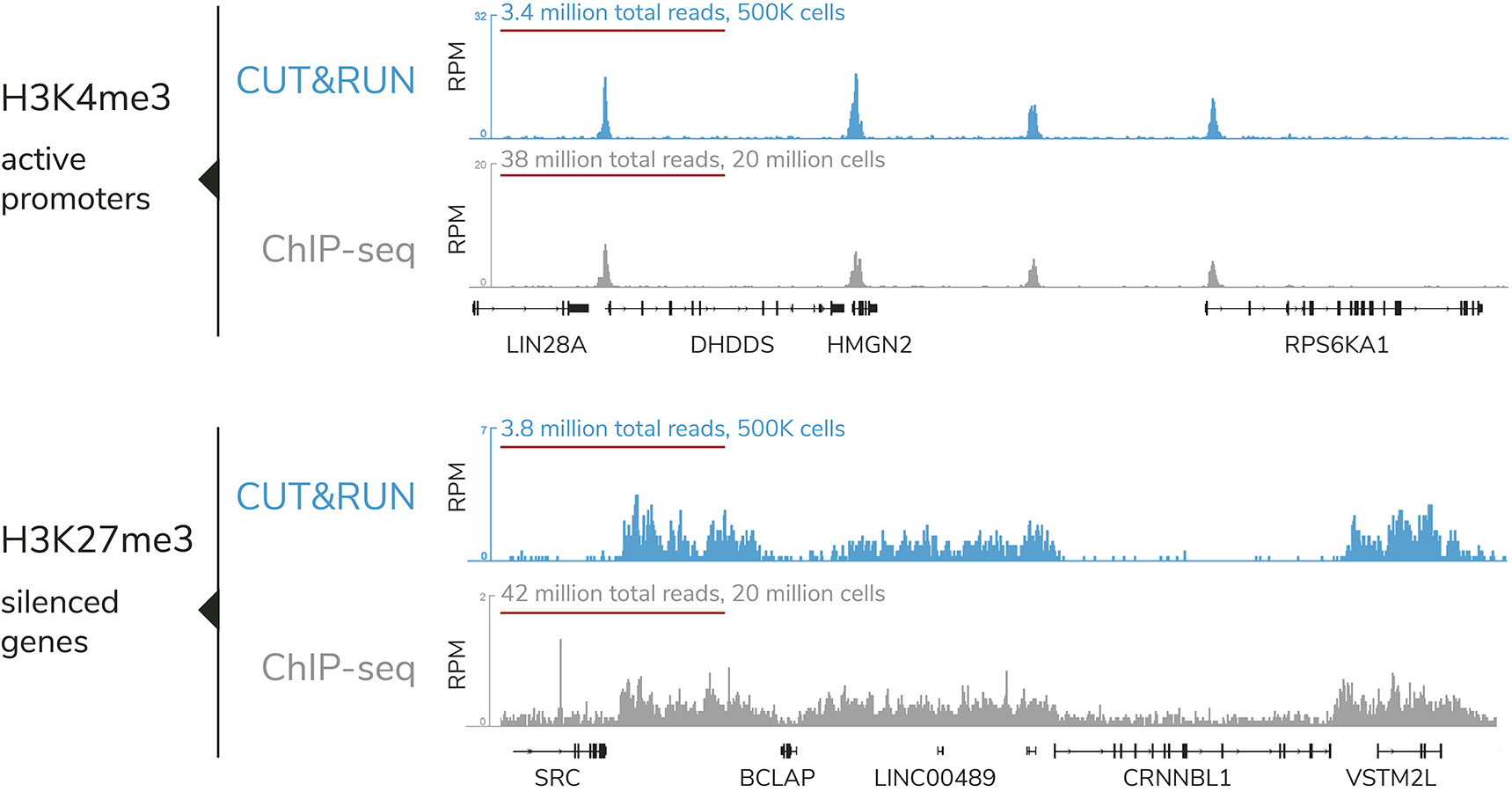
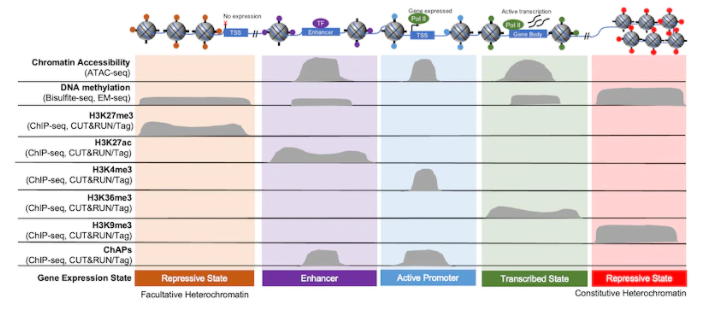
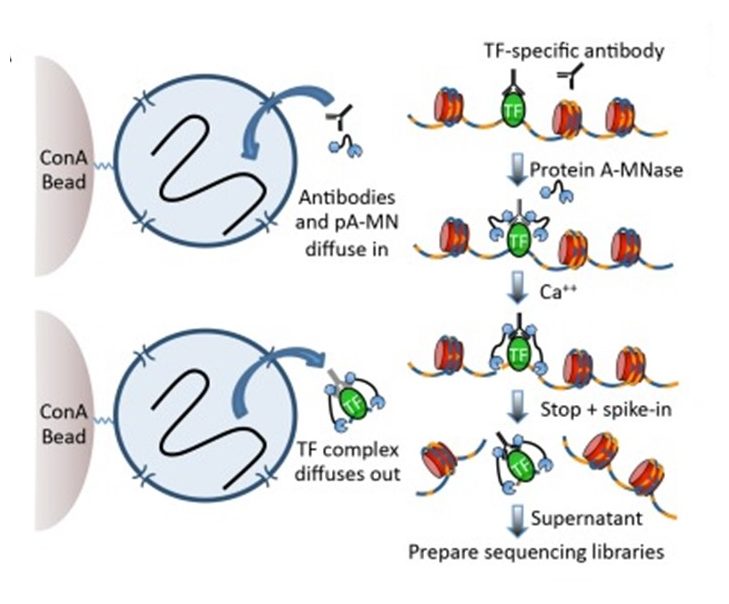
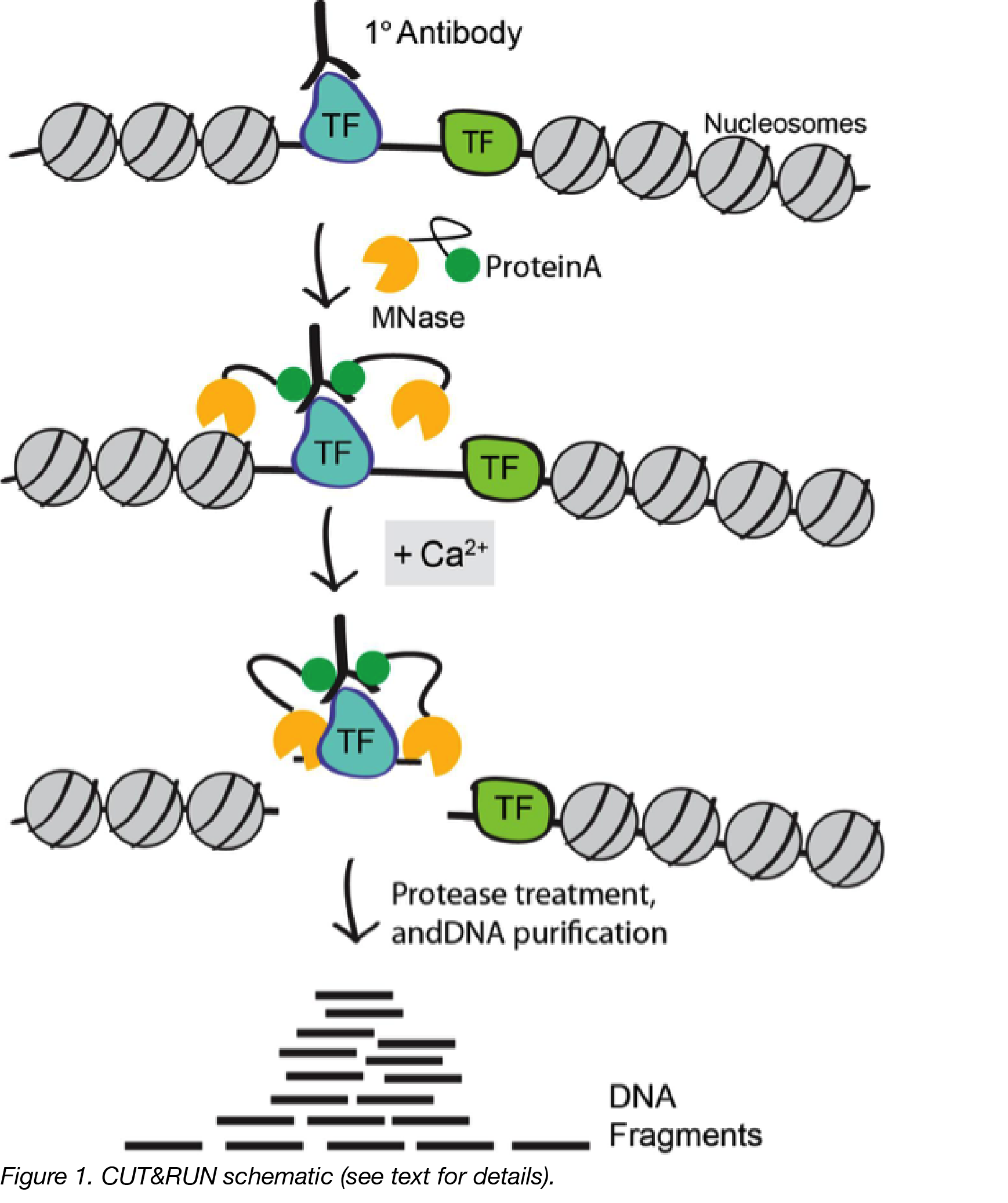
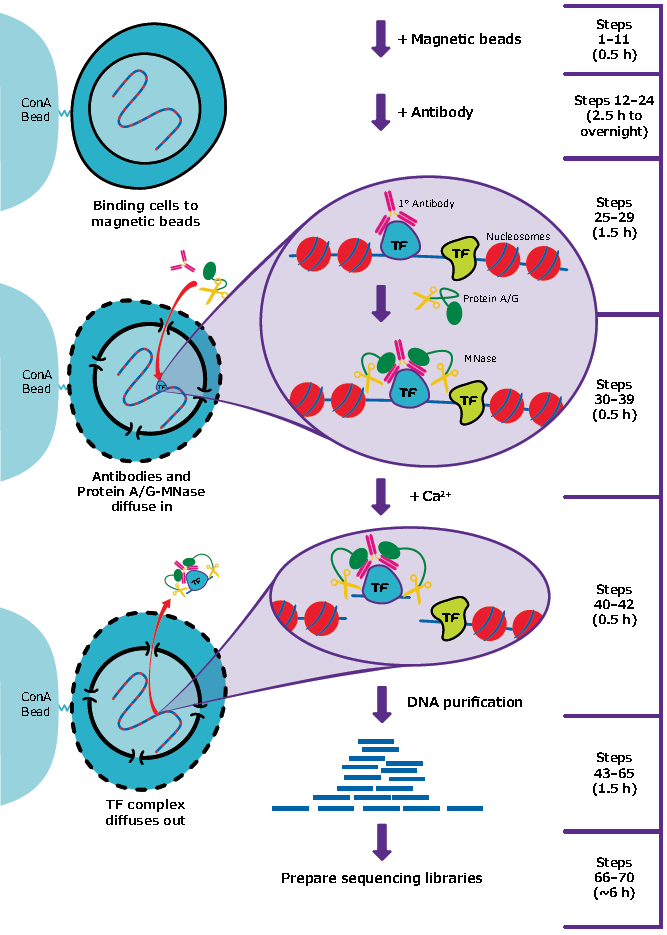
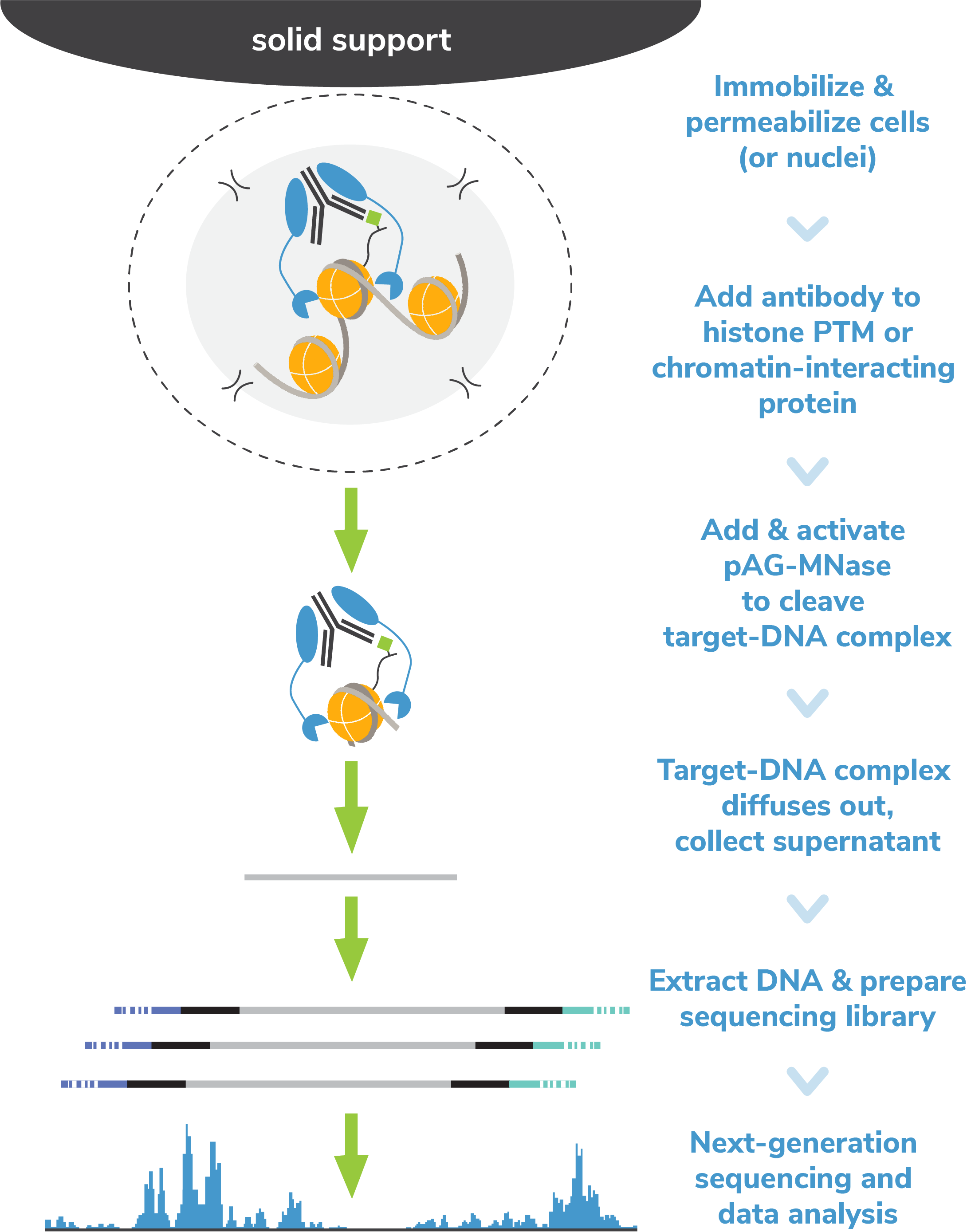
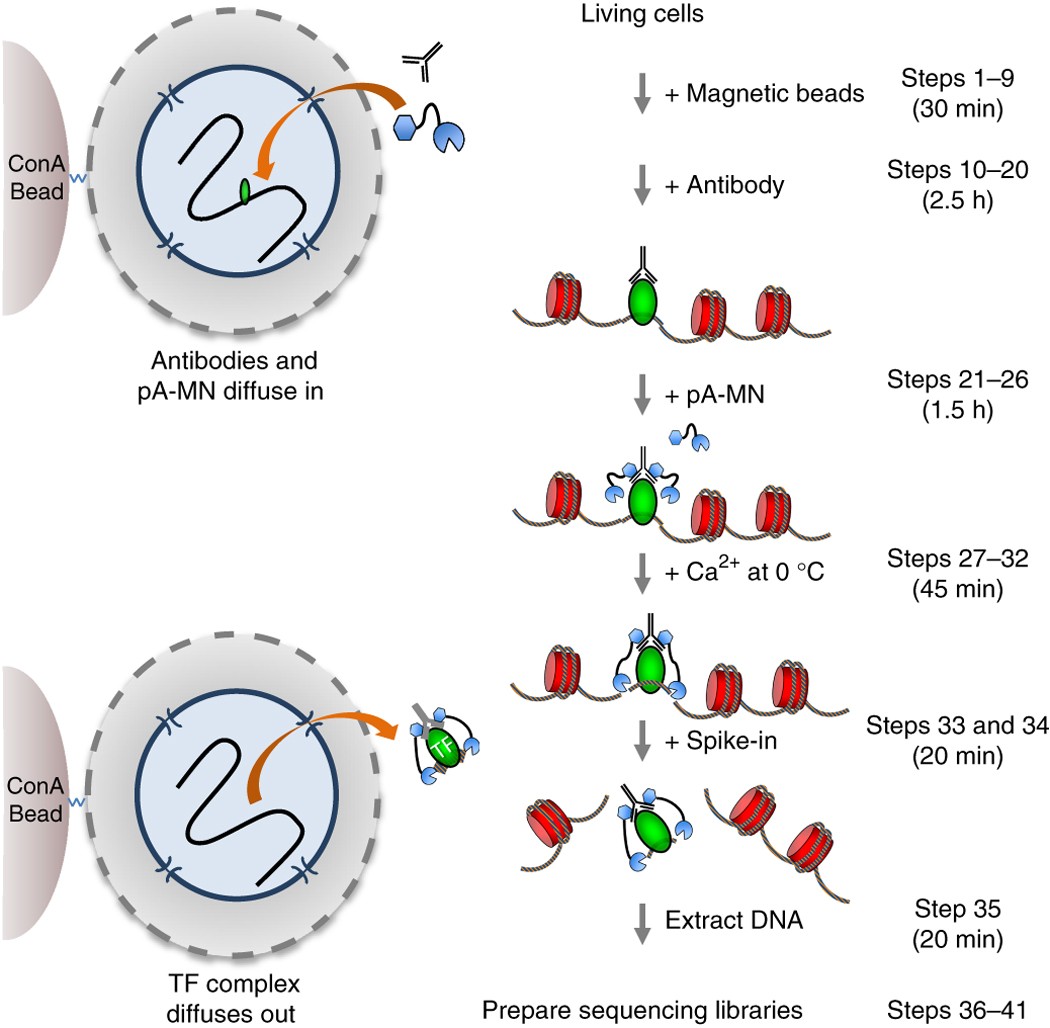
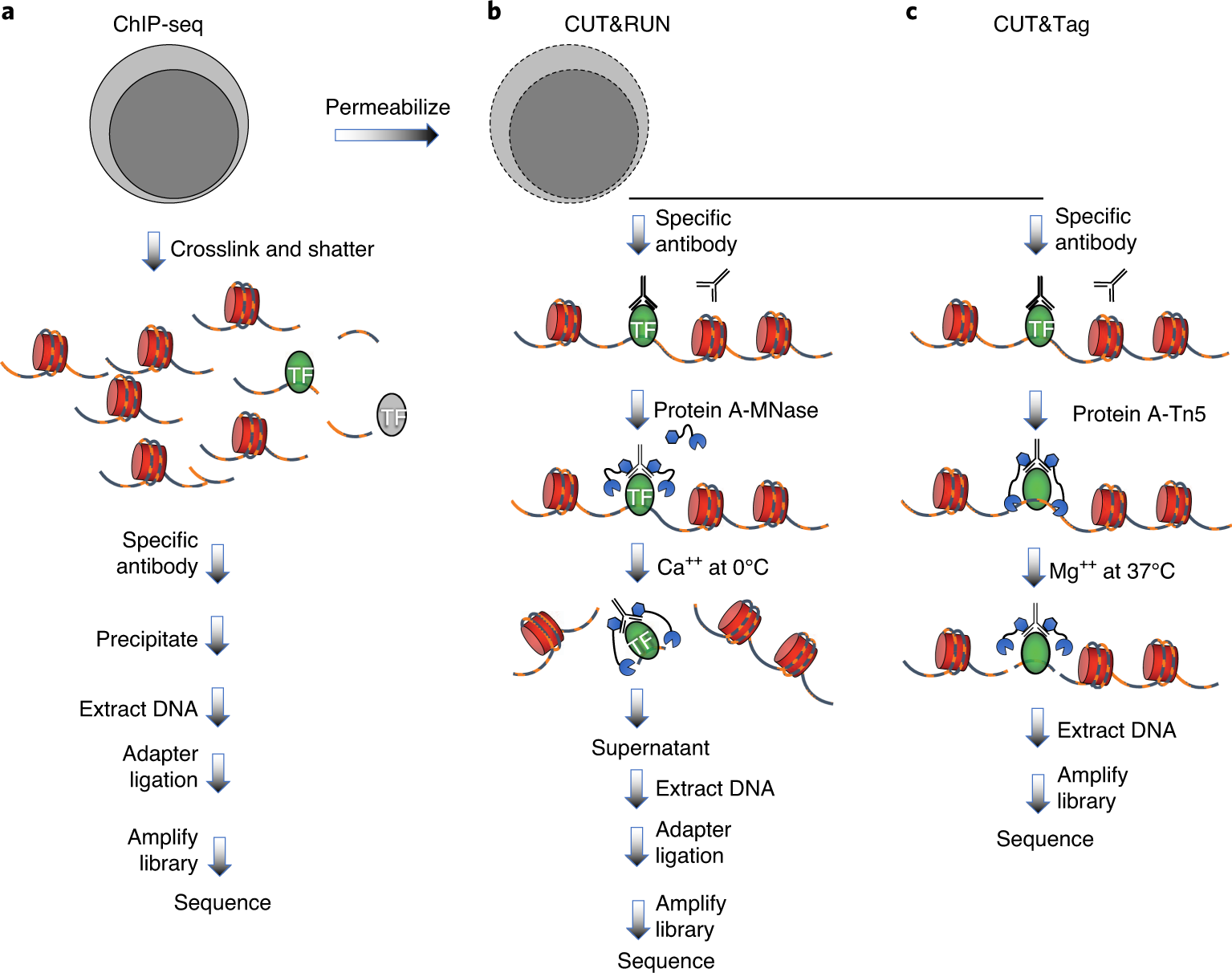
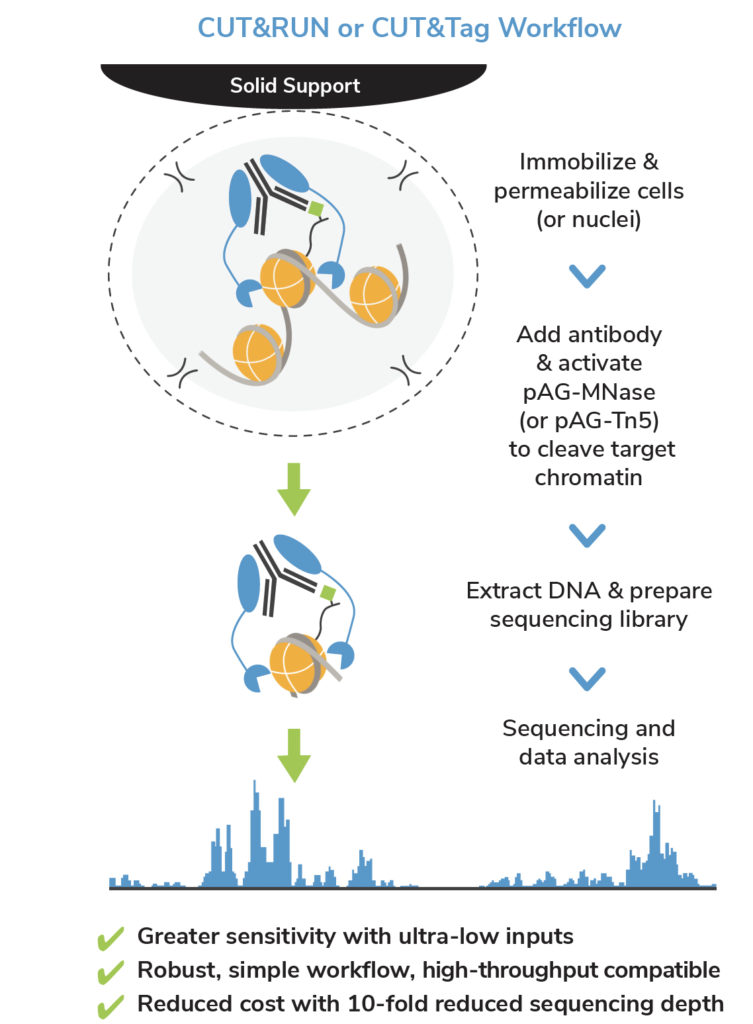
A modified CUT and RUN protocol and analysis pipeline to identify transcription factor binding sites in human cell lines: STAR Protocols
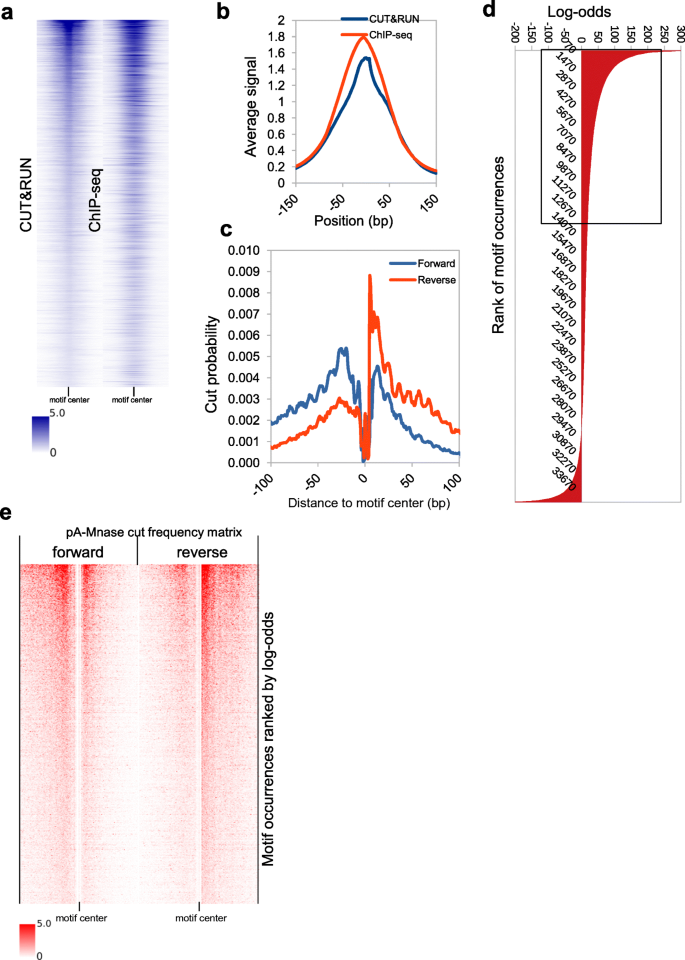
CUT&RUNTools: a flexible pipeline for CUT&RUN processing and footprint analysis | Genome Biology | Full Text

ChIP-seq vs CUT&RUN : Learn the benefits of CUT&RUN sequencing for enhanced chromatin profiling - YouTube

![Recombinant Anti-CTCF antibody [EPR18253] - ChIP Grade (ab188408) | Abcam Recombinant Anti-CTCF antibody [EPR18253] - ChIP Grade (ab188408) | Abcam](https://www.abcam.com/ps/products/188/ab188408/Images/ab188408-481303-anti-ctcf-antibody-epr18253-chip-grade-1.jpg)